What cluster analysis can do for you.
For the rest of the my explanation of cluster analysis I will be using the question I asked as an example (feel free to e-mail me if you're wondering how your data might fit).
My question:
Q1: Is the entire set of EGFP labeled somatostatin positive interneurons homogeneous in terms of their electrophysiological/pharmacological properties (such as action potential half-width, input resistance,firing rate accomodation, PSC sensitivity to drugs, etc.)?
Given that the EGFP labeled interneurons are only a subset of somatostatin positive interneurons - I could also ask:
Q2: Do these cells represent a distinct subset of SST+ve interneurons or are they a microcosm of SST+ve interneurons (i.e. How different are they from non-EGFP labeled SST+ve interneurons)?
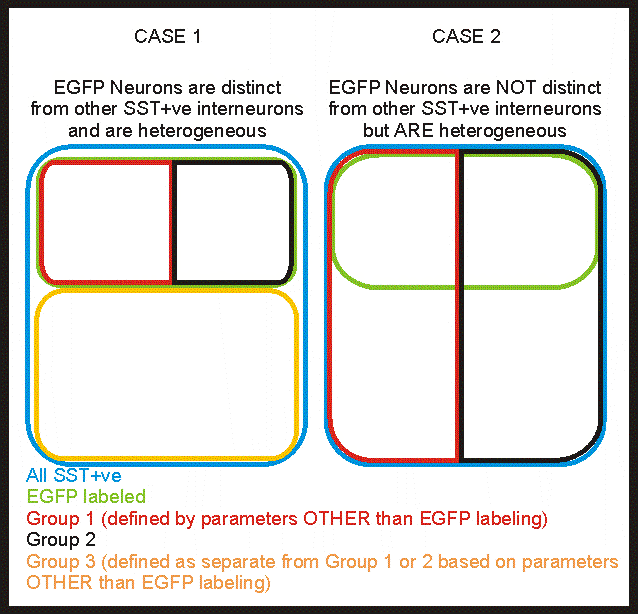
Perhaps these possible cases are better represented by these Venn diagrams:

From these you can tell how much data I need to get to answer my question.
Q1: Are EGFP labeled cells heterogeneous?
This is going be a discrimination between the Cases in the top box (1 & 2; where there are 2 groups within the 1 EGFP group) and the Cases in the bottom box (3 & 2; where there is a single group within the EGFP group)
Should I include all SST+ve interneurons in the dataset to answer this question?
No - it's not necessary. Adding non-EGFP labeled SST+ve interneurons will just allow discrimination between Cases 1 & 2 or Cases 3 & 4. Which leads to answering the next question...
Q2: Are EGFP cells different from the rest of the SST+ve interneurons?
The data recorded from all SST+ve interneurons will allow discrimination within the EGFP labeled group and discrimination between the EGFP labeled and non-EGFP labeled interneurons.
I'm going to skip ahead a bit here and show some tree diagrams (dendrograms) which is a common graphical representation of data after performing cluster analysis. Think of a taxonomic tree showing the current administration being more closely related to other primates than the rest of us.
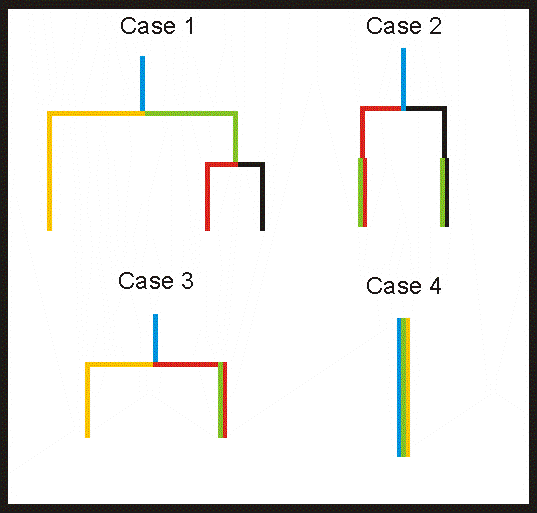
Description of these cases:
Case 1: The dataset combining all of the SST+ve interneurons can be classified into 2 groups based on electrophysiology (1 of which happens to also be labeled by EGFP). One group can be further classified into 2 more groups.
Case 2: The dataset can be classified into 2 groups based on electrophysiology (if EGFP labeled is included these could be further classified).
Both case 1 and 2 end up with more than 1 group of EGFP labeled interneurons (which would indicate that they are heterogeneous)
Case 3: The dataset can be classified into 2 groups based on electrophysiology (only one of which also overlaps with EGFP labeling)
Case 4: The dataset cannot be classified into any groups (unless EGFP labeling is used to discriminate)